16 Ex: Logistic - Maternal Risk (Hoffman)
Maternal Risk Factor for Low Birth Weight Delivery
Compiled: October 15, 2025
16.1 PREPARATION
16.1.2 Load Data
More complex example demonstrating modeling decisions
Another set of data from a study investigating predictors of low birth weight
idinfant’s unique identification number
Dependent variable (DV) or outcome
lowLow birth weight (outcome)0 = birth weight >2500 g (normal)
1 = birth weight < 2500 g (low))
bwtactual infant birth weight in grams (ignore for now)
Independent variables (IV) or predictors
ageAge of mother, in yearslwtMother’s weight at last menstrual period, in poundsftvNumber of physician visits in 1st trimester: 0 = None, 1 = One, … 6 = sixraceRace: 1 = White, 2 = Black, 3 = OtherptlHistory of premature labor: 0 = None, 1 = One, 2 = two, 3 = threehtHistory of hypertension: 1 = Yes, 0 = NosmokeSmoking status during pregnancy:1 = Yes, 0 = NouiUterine irritability: 1 = Yes, 0 = No
The data is saved in a text file (.txt) without any labels.
df_txt <- read.table("https://raw.githubusercontent.com/CEHS-research/data/master/Regression/lowbwt.txt",
header = TRUE,
sep = "",
na.strings = "NA",
dec = ".",
strip.white = TRUE)
tibble::glimpse(df_txt)Rows: 189
Columns: 11
$ id <int> 85, 86, 87, 88, 89, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101…
$ low <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ age <int> 19, 33, 20, 21, 18, 21, 22, 17, 29, 26, 19, 19, 22, 30, 18, 18, …
$ lwt <int> 182, 155, 105, 108, 107, 124, 118, 103, 123, 113, 95, 150, 95, 1…
$ race <int> 2, 3, 1, 1, 1, 3, 1, 3, 1, 1, 3, 3, 3, 3, 1, 1, 2, 1, 3, 1, 3, 1…
$ smoke <int> 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 1, 0, 1, 0, 0…
$ ptl <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0…
$ ht <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ ui <int> 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1…
$ ftv <int> 0, 3, 1, 2, 0, 0, 1, 1, 1, 0, 0, 1, 0, 2, 0, 0, 0, 3, 0, 1, 2, 3…
$ bwt <int> 2523, 2551, 2557, 2594, 2600, 2622, 2637, 2637, 2663, 2665, 2722…16.1.3 Wrangle Data
df_mom <- df_txt %>%
dplyr::mutate(id = factor(id)) %>%
dplyr::mutate(low = low %>%
factor() %>%
forcats::fct_recode("birth weight >2500 g (normal)" = "0",
"birth weight < 2500 g (low)" = "1")) %>%
dplyr::mutate(race = race %>%
factor() %>%
forcats::fct_recode("White" = "1",
"Black" = "2",
"Other" = "3")) %>%
dplyr::mutate(ptl_any = as.numeric(ptl > 0)) %>% # collapse into 0 = none vs. 1 = at least one
dplyr::mutate(ptl = factor(ptl)) %>% # declare the number of pre-term labors to be a factor: 0, 1, 2, 3
dplyr::mutate_at(vars(smoke, ht, ui, ptl_any), # declare all there variables to be factors with the same two levels
factor,
levels = 0:1,
labels = c("No", "Yes")) Display the structure of the ‘clean’ version of the dataset
'data.frame': 189 obs. of 12 variables:
$ id : Factor w/ 189 levels "4","10","11",..: 60 61 62 63 64 65 66 67 68 69 ...
$ low : Factor w/ 2 levels "birth weight >2500 g (normal)",..: 1 1 1 1 1 1 1 1 1 1 ...
$ age : int 19 33 20 21 18 21 22 17 29 26 ...
$ lwt : int 182 155 105 108 107 124 118 103 123 113 ...
$ race : Factor w/ 3 levels "White","Black",..: 2 3 1 1 1 3 1 3 1 1 ...
$ smoke : Factor w/ 2 levels "No","Yes": 1 1 2 2 2 1 1 1 2 2 ...
$ ptl : Factor w/ 4 levels "0","1","2","3": 1 1 1 1 1 1 1 1 1 1 ...
$ ht : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...
$ ui : Factor w/ 2 levels "No","Yes": 2 1 1 2 2 1 1 1 1 1 ...
$ ftv : int 0 3 1 2 0 0 1 1 1 0 ...
$ bwt : int 2523 2551 2557 2594 2600 2622 2637 2637 2663 2665 ...
$ ptl_any: Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...Rows: 189
Columns: 12
$ id <fct> 85, 86, 87, 88, 89, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 1…
$ low <fct> birth weight >2500 g (normal), birth weight >2500 g (normal), …
$ age <int> 19, 33, 20, 21, 18, 21, 22, 17, 29, 26, 19, 19, 22, 30, 18, 18…
$ lwt <int> 182, 155, 105, 108, 107, 124, 118, 103, 123, 113, 95, 150, 95,…
$ race <fct> Black, Other, White, White, White, Other, White, Other, White,…
$ smoke <fct> No, No, Yes, Yes, Yes, No, No, No, Yes, Yes, No, No, No, No, Y…
$ ptl <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0,…
$ ht <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes, No, No, N…
$ ui <fct> Yes, No, No, Yes, Yes, No, No, No, No, No, No, No, No, Yes, No…
$ ftv <int> 0, 3, 1, 2, 0, 0, 1, 1, 1, 0, 0, 1, 0, 2, 0, 0, 0, 3, 0, 1, 2,…
$ bwt <int> 2523, 2551, 2557, 2594, 2600, 2622, 2637, 2637, 2663, 2665, 27…
$ ptl_any <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, Yes, No, N…16.2 EXPLORATORY DATA ANALYSIS
16.2.1 Missing Data
df_mom %>%
dplyr::select("Race" = race,
"Low Birth Weight Delivery" = low,
"Age, years" = age,
"Weight, pounds" = lwt,
"1st Tri Dr Visits" = ftv,
"History of Premature Labor, any" = ptl_any,
"History of Premature Labor, number" = ptl,
"Smoking During pregnancy" = smoke,
"History of Hypertension" = ht,
"Uterince Irritability" = ui) %>%
naniar::miss_var_summary() %>%
dplyr::select(Variable = variable,
n = n_miss) %>%
flextable::flextable() %>%
apaSupp::theme_apa(caption = "Missing Data by Variable")Variable | n |
|---|---|
Race | 0 |
Low Birth Weight Delivery | 0 |
Age, years | 0 |
Weight, pounds | 0 |
1st Tri Dr Visits | 0 |
History of Premature Labor, any | 0 |
History of Premature Labor, number | 0 |
Smoking During pregnancy | 0 |
History of Hypertension | 0 |
Uterince Irritability | 0 |
16.2.2 Summary
df_mom %>%
dplyr::select("Low Birth Weight Delivery" = low,
"Race" = race,
"Age, yrs" = age,
"Weight, lbs" = lwt,
"1st Tri Dr Visits" = ftv,
"Hx of Premature Labor, number" = ptl,
"Hx Premature Labor, any" = ptl_any,
"Hx of Hypertension" = ht,
"Smoking During pregnancy" = smoke,
"Uterince Irritability" = ui) %>%
apaSupp::tab_freq(caption = "Summary of Categorical Variables")Statistic | ||
|---|---|---|
Low Birth Weight Delivery | ||
birth weight >2500 g (normal) | 130 (68.8%) | |
birth weight < 2500 g (low) | 59 (31.2%) | |
Race | ||
White | 96 (50.8%) | |
Black | 26 (13.8%) | |
Other | 67 (35.4%) | |
Hx of Premature Labor, number | ||
0 | 159 (84.1%) | |
1 | 24 (12.7%) | |
2 | 5 (2.6%) | |
3 | 1 (0.5%) | |
Hx Premature Labor, any | ||
No | 159 (84.1%) | |
Yes | 30 (15.9%) | |
Hx of Hypertension | ||
No | 177 (93.7%) | |
Yes | 12 (6.3%) | |
Smoking During pregnancy | ||
No | 115 (60.8%) | |
Yes | 74 (39.2%) | |
Uterince Irritability | ||
No | 161 (85.2%) | |
Yes | 28 (14.8%) | |
df_mom %>%
dplyr::select("Low Birth Weight Delivery" = low,
"Race" = race,
"Age, yrs" = age,
"Weight, lbs" = lwt,
"1st Tri Dr Visits" = ftv,
"Hx Premature Labor, any" = ptl_any,
"Hx Premature Labor, number" = ptl,
"Hx Hypertension" = ht,
"Smoking During pregnancy" = smoke,
"Uterince Irritability" = ui) %>%
apaSupp::tab_desc(caption = "Summary of Continuous Variables")NA | M | SD | min | Q1 | Mdn | Q3 | max | |
|---|---|---|---|---|---|---|---|---|
Age, yrs | 0 | 23.24 | 5.30 | 14 | 19.00 | 23 | 26.00 | 45 |
Weight, lbs | 0 | 129.81 | 30.58 | 80 | 110.00 | 121 | 140.00 | 250 |
1st Tri Dr Visits | 0 | 0.79 | 1.06 | 0 | 0.00 | 0 | 1.00 | 6 |
Note. N = 189. NA = not available or missing; Mdn = median; Q1 = 25th percentile; Q3 = 75th percentile. | ||||||||
16.3 UNADJUSTED
Unadjusted Models
16.3.1 Fit Models
fit_glm_race <- glm(low ~ race, family = binomial(link = "logit"), data = df_mom)
fit_glm_age <- glm(low ~ age, family = binomial(link = "logit"), data = df_mom)
fit_glm_lwt <- glm(low ~ lwt, family = binomial(link = "logit"), data = df_mom)
fit_glm_ftv <- glm(low ~ ftv, family = binomial(link = "logit"), data = df_mom)
fit_glm_ptl <- glm(low ~ ptl_any, family = binomial(link = "logit"), data = df_mom)
fit_glm_ht <- glm(low ~ ht, family = binomial(link = "logit"), data = df_mom)
fit_glm_smoke <- glm(low ~ smoke, family = binomial(link = "logit"), data = df_mom)
fit_glm_ui <- glm(low ~ ui, family = binomial(link = "logit"), data = df_mom)16.3.2 Parameter Tables
Note: the parameter estimates here are for the LOGIT scale, not the odds ration (OR) or even the probability.
apaSupp::tab_glms(list(fit_glm_race, fit_glm_age, fit_glm_lwt, fit_glm_ftv),
narrow = TRUE,
fit = NA,
pr2 = "tjur")
| Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI |
race | ||||||||
White | — | — | ||||||
Black | 2.33 | [0.93, 5.77] | ||||||
Other | 1.89 | [0.96, 3.76] | ||||||
age | 0.95 | [0.89, 1.01] | ||||||
lwt | 0.99 | [0.97, 1.00]* | ||||||
ftv | 0.87 | [0.63, 1.17] | ||||||
pseudo-R² | .026 | .014 | .031 | .004 | ||||
Note. N = 189. CI = confidence interval.Coefficient of determination estiamted by Tjur's psuedo R-squared | ||||||||
* p < .05. ** p < .01. *** p < .001. | ||||||||
apaSupp::tab_glms(list(fit_glm_ptl, fit_glm_ht, fit_glm_smoke, fit_glm_ui),
narrow = TRUE,
fit = NA,
pr2 = "tjur")
| Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI |
ptl_any | ||||||||
No | — | — | ||||||
Yes | 4.32 | [1.94, 9.95]*** | ||||||
ht | ||||||||
No | — | — | ||||||
Yes | 3.37 | [1.03, 11.83]* | ||||||
smoke | ||||||||
No | — | — | ||||||
Yes | 2.02 | [1.08, 3.80]* | ||||||
ui | ||||||||
No | — | — | ||||||
Yes | 2.58 | [1.13, 5.88]* | ||||||
pseudo-R² | .073 | .023 | .026 | .029 | ||||
Note. N = 189. CI = confidence interval.Coefficient of determination estiamted by Tjur's psuedo R-squared | ||||||||
* p < .05. ** p < .01. *** p < .001. | ||||||||
16.4 MAIN EFFECTS ONLY
Main-effects multiple logistic regression model
fit_glm_mains <- glm(low ~ race + age + lwt + ftv + ptl_any + ht + smoke + ui,
family = binomial(link = "logit"),
data = df_mom)apaSupp::tab_glm(fit_glm_mains,
var_labels = c(race = "Race",
age = "Age, yrs",
lwt = "Prior Weight, lbs",
ftv = "First Tri Visits",
ptl_any = "Hx Premature Labor",
ht = "Hx Hypertension",
smoke = "Smoking",
ui = "Uterine Irritability"),
show_single_row = c("ptl_any", "smoke", "ht", "ui"),
vif = FALSE) %>%
flextable::width(j = 1, width = 1.75)Odds Ratio | Logit Scale | |||||||
|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | Wald | LRT | ||
Race | .039* | |||||||
White | — | — | — | — | ||||
Black | 3.38 | [1.19, 9.81] | 1.219 | (0.53) | .022* | |||
Other | 2.27 | [0.95, 5.60] | .819 | (0.45) | .069 | |||
Age, yrs | 0.96 | [0.89, 1.03] | -0.04 | (0.04) | .302 | .297 | ||
Prior Weight, lbs | 0.99 | [0.97, 1.00] | -0.02 | (0.01) | .032* | .024* | ||
First Tri Visits | 1.05 | [0.74, 1.48] | 0.05 | (0.18) | .772 | .772 | ||
Hx Premature Labor | 3.38 | [1.38, 8.57] | 1.22 | (0.46) | .008** | .008** | ||
Hx Hypertension | 6.43 | [1.66, 28.19] | 1.86 | (0.71) | .009** | .007** | ||
Smoking | 2.36 | [1.07, 5.38] | 0.86 | (0.41) | .036* | .034* | ||
Uterine Irritability | 2.05 | [0.82, 5.10] | 0.72 | (0.46) | .121 | .124 | ||
(Intercept) | 0.64 | (1.22) | .598 | |||||
pseudo-R² | .191 | |||||||
Note. N = 189. CI = confidence interval. Significance denotes Wald t-tests for individual parameter estimates, as well as Likelihood Ratio Tests (LRT) for single-predictor deletion. Coefficient of determination displays Tjur's pseudo-R². | ||||||||
* p < .05. ** p < .01. *** p < .001. | ||||||||
16.5 INTERACTIONS
Before removing non-significant main effects, test plausible interactions
Try the following interactions:
Age and Weight
Age and Smoking
Weight and Smoking
fit_glm_mains_aw <- glm(low ~ race + age + lwt + ftv + ptl_any + ht + smoke + ui + age:lwt,
family = binomial(link = "logit"),
data = df_mom)
fit_glm_mains_as <- glm(low ~ race + age + lwt + ftv + ptl_any + ht + smoke + ui + age:smoke,
family = binomial(link = "logit"),
data = df_mom)
fit_glm_mains_ws <- glm(low ~ race + age + lwt + ftv + ptl_any + ht + smoke + ui + lwt:smoke,
family = binomial(link = "logit"),
data = df_mom)apaSupp::tab_glms(list("Age-Weight" = fit_glm_mains_aw,
"Age-Smoking" = fit_glm_mains_as,
"Weight-Smoking" = fit_glm_mains_ws),
var_labels = c(race = "Race",
age = "Age, yrs",
lwt = "Prior Weight, lbs",
ftv = "First Tri Visits",
ptl_any = "Hx Premature Labor",
smoke = "Smoking",
ht = "Hypertension",
ui = "Uterine Irritability"),
narrow = TRUE,
show_single_row = c("ptl_any", "smoke", "ht", "ui"))
| Age-Weight | Age-Smoking | Weight-Smoking | |||
|---|---|---|---|---|---|---|
Variable | OR | 95% CI | OR | 95% CI | OR | 95% CI |
Race | ||||||
White | — | — | — | — | — | — |
Black | 3.36 | [1.18, 9.78]* | 3.10 | [1.07, 9.18]* | 3.44 | [1.21, 9.99]* |
Other | 2.25 | [0.94, 5.60] | 2.18 | [0.90, 5.41] | 2.06 | [0.85, 5.13] |
Age, yrs | 0.94 | [0.67, 1.33] | 0.93 | [0.83, 1.03] | 0.96 | [0.89, 1.04] |
Prior Weight, lbs | 0.98 | [0.92, 1.04] | 0.99 | [0.97, 1.00]* | 0.98 | [0.95, 1.00]* |
First Tri Visits | 1.05 | [0.74, 1.48] | 1.04 | [0.73, 1.46] | 1.03 | [0.72, 1.44] |
Hx Premature Labor | 3.40 | [1.38, 8.64]** | 3.30 | [1.35, 8.34]** | 3.54 | [1.44, 9.01]** |
Hypertension | 6.43 | [1.66, 28.23]** | 6.54 | [1.69, 28.82]** | 5.79 | [1.48, 25.60]* |
Smoking | 2.35 | [1.06, 5.36]* | 0.57 | [0.02, 18.96] | 0.33 | [0.01, 10.04] |
Uterine Irritability | 2.06 | [0.82, 5.13] | 2.20 | [0.86, 5.59] | 2.23 | [0.88, 5.64] |
age * lwt | 1.00 | [1.00, 1.00] | ||||
age * smoke | ||||||
age * Yes | 1.06 | [0.92, 1.24] | ||||
lwt * smoke | ||||||
lwt * Yes | 1.02 | [0.99, 1.04] | ||||
Fit Metrics | ||||||
AIC | 218.7 | 218.1 | 217.4 | |||
BIC | 254.4 | 253.7 | 253.1 | |||
pseudo-R² | ||||||
Tjur | .191 | .192 | .196 | |||
McFadden | .162 | .164 | .167 | |||
Note. N = 189. CI = confidence interval.Coefficient of determination estiamted with both Tjur and McFadden's psuedo R-squared | ||||||
* p < .05. ** p < .01. *** p < .001. | ||||||
apaSupp::tab_glm_fits(list("Only Mains" = fit_glm_mains,
"Age-Weight" = fit_glm_mains_aw,
"Age-Smoking" = fit_glm_mains_as,
"Weight-Smoking" = fit_glm_mains_ws))pseudo- | |||||||
|---|---|---|---|---|---|---|---|
Model | N | k | McFadden | Tjur | AIC | BIC | RMSE |
Only Mains | 189 | 10 | .162 | .191 | 216.75 | 249.17 | 0.42 |
Age-Weight | 189 | 11 | .162 | .191 | 218.73 | 254.39 | 0.42 |
Age-Smoking | 189 | 11 | .164 | .192 | 218.08 | 253.74 | 0.42 |
Weight-Smoking | 189 | 11 | .167 | .196 | 217.42 | 253.08 | 0.42 |
Note. k = number of parameters estimated in each model. Larger values indicated better performance. Smaller values indicated better performance for Akaike's Information Criteria (AIC), Bayesian information criteria (BIC), and Root Mean Squared Error (RMSE). | |||||||
# A tibble: 2 × 5
`Resid. Df` `Resid. Dev` Df Deviance `Pr(>Chi)`
<dbl> <dbl> <dbl> <dbl> <dbl>
1 179 197. NA NA NA
2 178 197. 1 0.0190 0.890# A tibble: 2 × 5
`Resid. Df` `Resid. Dev` Df Deviance `Pr(>Chi)`
<dbl> <dbl> <dbl> <dbl> <dbl>
1 179 197. NA NA NA
2 178 196. 1 0.669 0.413# A tibble: 2 × 5
`Resid. Df` `Resid. Dev` Df Deviance `Pr(>Chi)`
<dbl> <dbl> <dbl> <dbl> <dbl>
1 179 197. NA NA NA
2 178 195. 1 1.33 0.24916.6 PARSAMONY
No interactions are significant Remove non-significant main effects
Since the mother’s age is theoretically a meaningful variable, it should probably be retained.
Remove “UI” since its not significant
fit_glm_trim <- glm(low ~ race + age + lwt + ptl_any + ht + smoke ,
family = binomial(link = "logit"),
data = df_mom)apaSupp::tab_glm(fit_glm_trim,
var_labels = c(race = "Race",
age = "Age, yrs",
lwt = "Prior Weight, lbs",
ptl_any = "Hx Premature Labor",
ht = "Hx Hypertension",
smoke = "Smoking"),
show_single_row = c("ptl_any", "smoke", "ht"),
lrt = FALSE,
vif = FALSE) %>%
flextable::width(j = 1, width = 1.75)Odds Ratio | Logit Scale | ||||||
|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | p | ||
Race | |||||||
White | — | — | — | — | |||
Black | 3.22 | [1.13, 9.31] | 1.168 | (0.53) | .028* | ||
Other | 2.26 | [0.96, 5.50] | .815 | (0.44) | .066 | ||
Age, yrs | 0.96 | [0.89, 1.03] | -0.04 | (0.04) | .255 | ||
Prior Weight, lbs | 0.98 | [0.97, 1.00] | -0.02 | (0.01) | .028* | ||
Hx Premature Labor | 3.80 | [1.57, 9.53] | 1.33 | (0.46) | .004** | ||
Hx Hypertension | 5.70 | [1.49, 24.76] | 1.74 | (0.70) | .013* | ||
Smoking | 2.36 | [1.08, 5.32] | 0.86 | (0.40) | .034* | ||
(Intercept) | 0.92 | (1.20) | .442 | ||||
pseudo-R² | .180 | ||||||
Note. N = 189. CI = confidence interval. Significance denotes Wald t-tests for parameter estimates. Coefficient of determination displays Tjur's pseudo-R². | |||||||
* p < .05. ** p < .01. *** p < .001. | |||||||
16.7 CENTER & SCALE
Since the mother’s age is theoretically a meaningful variable, it should probably be retained.
Revise so that age is interpreted in 5-year and pre-pregnancy weight in 20 lb increments and the intercept has meaning.
fit_glm_final <- glm(low ~ race + I((age - 20)/5) + I((lwt - 125)/20)
+ ptl_any + ht + smoke ,
family = binomial(link = "logit"),
data = df_mom)apaSupp::tab_glm(fit_glm_final,
var_labels = c("I((age - 20)/5)" = "Age, 5 yr",
"I((lwt - 125)/20)" = "Pre-Preg Weight, 20 lb",
race = "Race",
smoke = "Smoker",
ptl_any = "Prior Preterm Labor",
ht = "Hx Hypertension"),
p_note = "apa13",
show_single_row = c("smoke", "ptl_any", "ht"),
general_note = "Centering for age (20 yr) and weight (125 lbs)")Odds Ratio | Logit Scale | ||||||||
|---|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | Wald | LRT | VIF | ||
Race | .044* | 1.49 | |||||||
White | — | — | — | — | |||||
Black | 3.22 | [1.13, 9.31] | 1.168 | (0.53) | .028* | ||||
Other | 2.26 | [0.96, 5.50] | .815 | (0.44) | .066 | ||||
Age, 5 yr | 0.81 | [0.55, 1.16] | -0.21 | (0.19) | .255 | .249 | 1.09 | ||
Pre-Preg Weight, 20 lb | 0.73 | [0.55, 0.95] | -0.31 | (0.14) | .028* | .020* | 1.29 | ||
Prior Preterm Labor | 3.80 | [1.57, 9.53] | 1.33 | (0.46) | .004** | .003** | 1.08 | ||
Hx Hypertension | 5.70 | [1.49, 24.76] | 1.74 | (0.70) | .013* | .011* | 1.15 | ||
Smoker | 2.36 | [1.08, 5.32] | 0.86 | (0.40) | .034* | .032* | 1.35 | ||
(Intercept) | -1.86 | (0.41) | < .001*** | ||||||
pseudo-R² | .180 | ||||||||
Note. N = 189. CI = confidence interval; VIF = variance inflation factor. Significance denotes Wald t-tests for individual parameter estimates, as well as Likelihood Ratio Tests (LRT) for single-predictor deletion. Coefficient of determination displays Tjur's pseudo-R². Centering for age (20 yr) and weight (125 lbs) | |||||||||
* p < .05. *** p < .001. | |||||||||
16.8 Probe & Plot
16.8.1 focus: Compare Races with prior weight and hypertension
interactions::interact_plot(model = fit_glm_final,
pred = lwt,
modx = race,
legend.main = "Race",
mod2 = ht,
mod2.labels = c("History of Hypertension: No",
"History of Hypertension: Yes")) +
theme_bw() +
theme(legend.position = "inside",
legend.position.inside = c(.5, 1),
legend.justification = c(1.1, 1.1),
legend.background = element_rect(color = "black"),
legend.key.width = unit(1.5, "cm")) +
scale_y_continuous(labels = scales::percent_format()) +
labs(x = "Mother's Weight Pre-Pregnancy, pounds",
y = "Predicted Probability\nBaby has Low Birth Weight (< 2500 grams)",
linetype = "Race") +
scale_linetype_manual(values = c("solid", "longdash", "dotted"))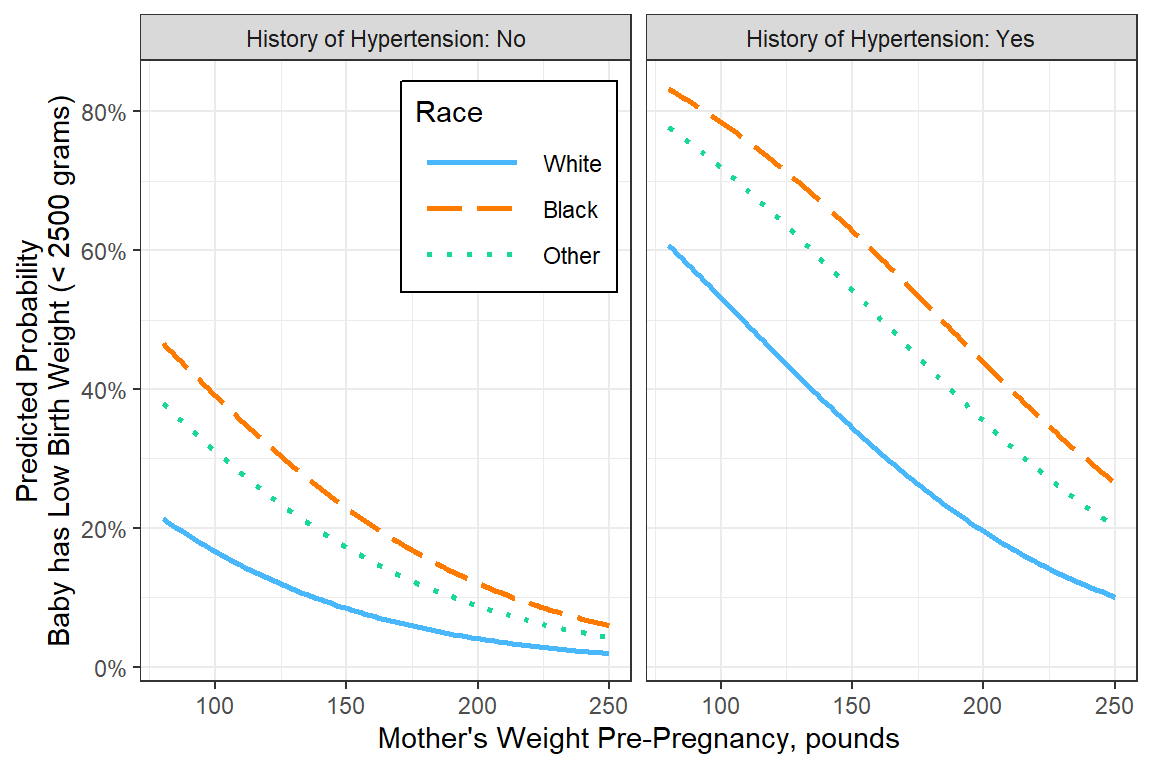
effects::Effect(mod = fit_glm_trim,
focal.predictors = c("lwt", "race", "ptl_any", "ht", "smoke"),
xlevels = list(lwt = seq(from = 80, to = 250, by = 5))) %>%
data.frame() %>%
dplyr::filter(race != "Other") %>%
dplyr::mutate(risk = interaction(ptl_any, ht, smoke) %>%
factor() %>%
forcats::fct_recode("(0) None" = "No.No.No",
"(1) Preterm" = "Yes.No.No" ,
"(1) Ht" = "No.Yes.No" ,
"(1) Smoking" = "No.No.Yes" ,
"(2) Preterm & Ht" = "Yes.Yes.No" ,
"(2) Ht & Smoking" = "No.Yes.Yes" ,
"(2) Preterm & Smoking" = "Yes.No.Yes",
"(3) All Three" = "Yes.Yes.Yes") %>%
forcats::fct_reorder(fit) %>%
forcats::fct_rev()) %>%
ggplot(aes(x = lwt,
y = fit)) +
geom_line(aes(color = risk,
linetype = risk,
linewidth = risk)) +
theme_bw() +
facet_grid(~ race) +
scale_linetype_manual(values = c("solid",
"longdash", "dotdash", "dashed",
"solid", "solid", "solid",
"solid")) +
scale_linewidth_manual(values = c(1.5,
1.25, 1.25, 1.25,
.75, .75, .75,
.5)) +
scale_color_manual(values = c("black",
"red", "coral", "orange",
"purple", "dodgerblue", "cyan3",
"green4")) +
scale_y_continuous(labels = scales::percent_format()) +
labs(x = "Mother's Weight Pre-Pregnancy, pounds",
y = "Predicted Probability\nBaby has Low Birth Weight (< 2500 grams)",
color = "Risk Factors:",
linetype = "Risk Factors:",
linewidth= "Risk Factors:") +
theme(#legend.position = "bottom",
legend.key.width = unit(1.5, "cm")) 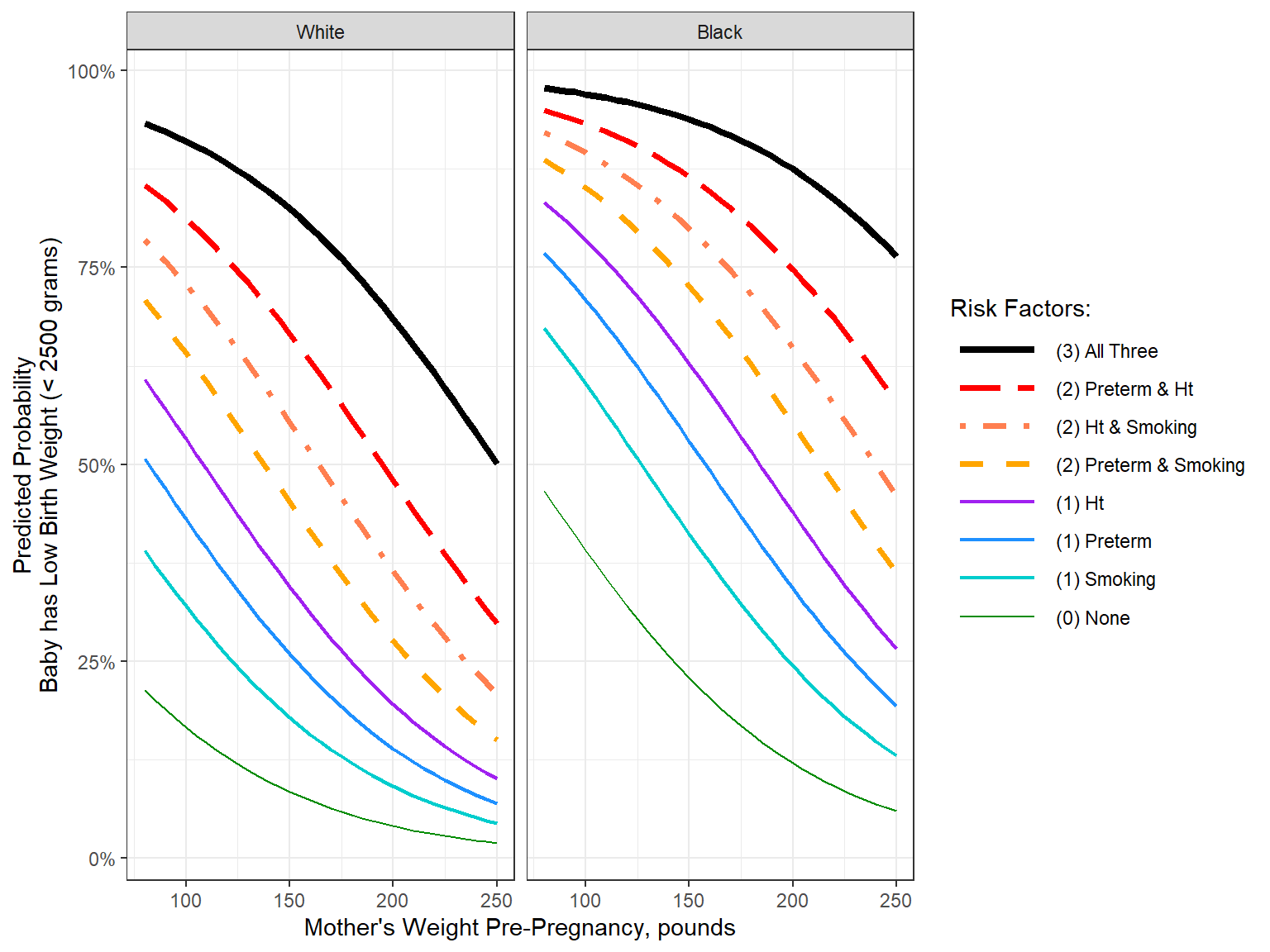
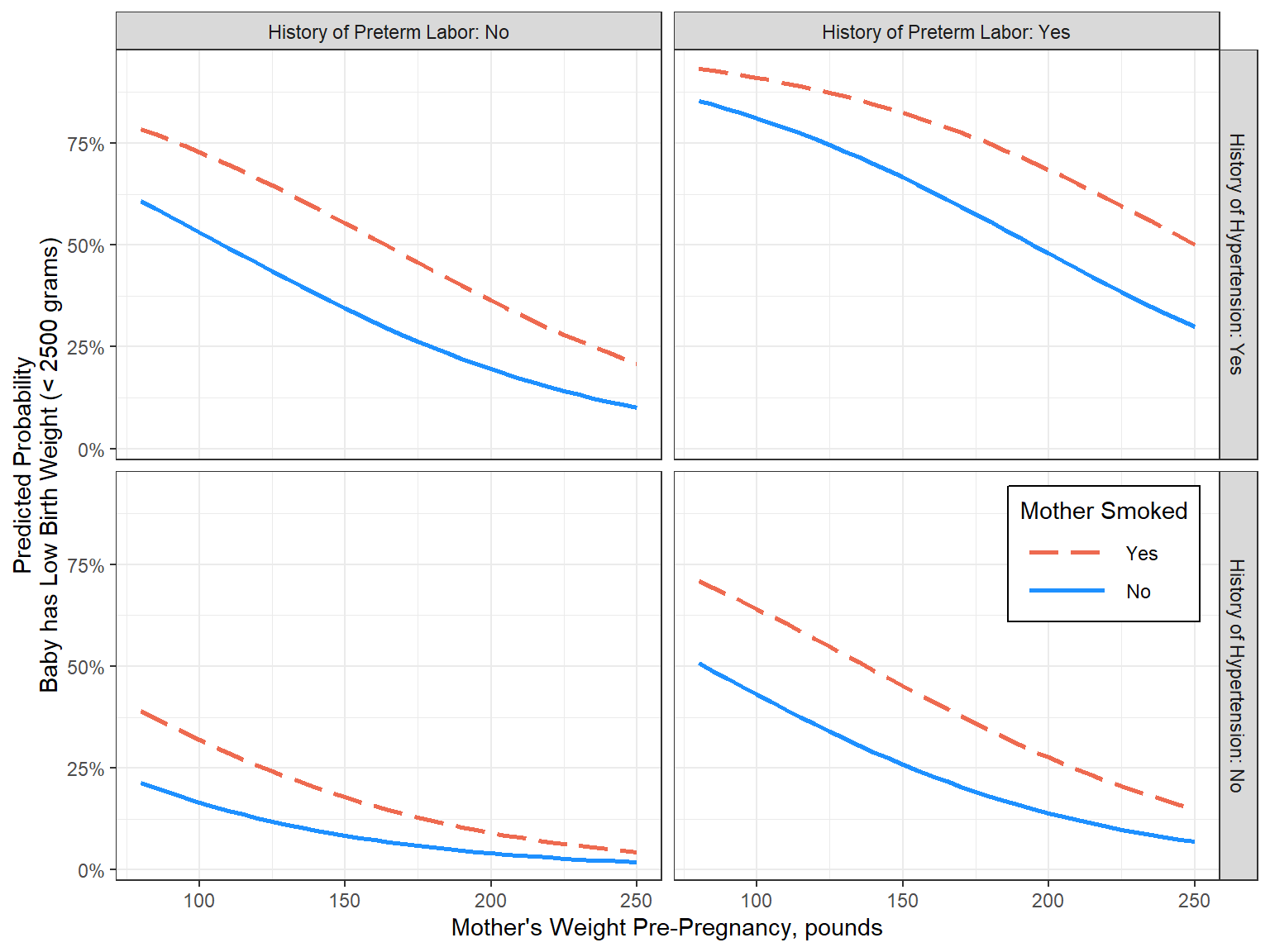
16.8.1.1 Focus on: Mother’s weight and smoking status during pregnancy, as well as history of any per-term labor and hypertension
Illustates risk given the mother is 20 years old and white
effects::Effect(mod = fit_glm_final,
focal.predictors = c("race", "lwt", "smoke", "ptl_any", "ht"),
fixed.predictors = list(age = 20),
xlevels = list(lwt = seq(from = 80, to = 250, by = 5))) %>%
data.frame() %>%
dplyr::filter(race == "White") %>%
dplyr::mutate(smoke = forcats::fct_rev(smoke)) %>%
dplyr::mutate(ptl_any_labels = glue::glue("History of Preterm Labor: {ptl_any}")) %>%
dplyr::mutate(ht_labels = glue::glue("History of Hypertension: {ht}") %>% forcats::fct_rev()) %>%
ggplot(aes(x = lwt,
y = fit)) +
geom_line(aes(color = smoke,
linetype = smoke),
linewidth = 1) +
theme_bw() +
facet_grid(ht_labels ~ ptl_any_labels) +
scale_y_continuous(labels = scales::percent_format()) +
labs(x = "Mother's Weight Pre-Pregnancy, pounds",
y = "Predicted Probability\nBaby has Low Birth Weight (< 2500 grams)",
color = "Mother Smoked",
linetype = "Mother Smoked") +
theme(legend.position = "inside",
legend.position.inside = c(1, .5),
legend.justification = c(1.1, 1.15),
legend.background = element_rect(color = "black"),
legend.key.width = unit(1.5, "cm")) +
scale_linetype_manual(values = c("longdash", "solid")) +
scale_color_manual(values = c( "coral2", "dodger blue"))