14 Ex: Logistic - Depression (Hoffman)
Compiled: October 15, 2025
14.1 PREPARATION
14.1.2 Load Data
This dataset comes from John Hoffman’s textbook: Regression Models for Categorical, Count, and Related Variables: An Applied Approach (2004) Amazon link, 2014 edition
Chapter 3: Logistic and Probit Regression Models
Dataset: The following example uses the SPSS data set Depress.sav. The dependent variable of interest is a measure of life satisfaction, labeled satlife.
Dependent Variable = satlife (numeric version) or lifesat (factor version)
df_depress <- haven::read_spss("https://raw.githubusercontent.com/CEHS-research/data/master/Hoffmann_datasets/depress.sav") %>%
haven::as_factor() %>% # labelled to factors
haven::zap_label() %>% # remove SPSS junk
haven::zap_formats() %>% # remove SPSS junk
haven::zap_widths() %>% # remove SPSS junk
dplyr::mutate(sex = forcats::fct_recode(sex,
"Female" = "female",
"Male" = "male")) %>%
dplyr::mutate(lifesat = forcats::fct_recode(lifesat,
"Yes (1)" = "high",
"No (0)" = "low"))Rows: 118
Columns: 14
$ id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,…
$ age <dbl> 39, 41, 42, 30, 35, 44, 31, 39, 35, 33, 38, 31, 40, 44, 43, 32…
$ iq <dbl> 94, 89, 83, 99, 94, 90, 94, 87, NA, 92, 92, 94, 91, 86, 90, NA…
$ anxiety <fct> medium low, medium low, medium high, medium low, medium low, N…
$ depress <fct> medium, medium, high, medium, low, low, medium, medium, medium…
$ sleep <fct> low, low, low, low, high, low, NA, low, low, low, high, low, l…
$ sex <fct> Female, Female, Female, Female, Female, Male, Female, Female, …
$ lifesat <fct> No (0), No (0), No (0), No (0), Yes (1), Yes (1), No (0), Yes …
$ weight <dbl> 4.9, 2.2, 4.0, -2.6, -0.3, 0.9, -1.5, 3.5, -1.2, 0.8, -1.9, 5.…
$ satlife <dbl> 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 0,…
$ male <dbl> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0,…
$ sleep1 <dbl> 0, 0, 0, 0, 1, 0, NA, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, N…
$ newiq <dbl> 2.21, -2.79, -8.79, 7.21, 2.21, -1.79, 2.21, -4.79, NA, 0.21, …
$ newage <dbl> 1.5424, 3.5424, 4.5424, -7.4576, -2.4576, 6.5424, -6.4576, 1.5…# A tibble: 3 × 3
lifesat satlife n
<fct> <dbl> <int>
1 Yes (1) 1 52
2 No (0) 0 65
3 <NA> NA 114.2 EXPLORATORY DATA ANALYSIS
14.2.1 Missing Data
df_depress %>%
dplyr::select("Sex" = sex,
"Life Satisfaction" = lifesat,
"IQ, pts" = iq,
"Age, yrs" = age,
"Weight, lbs" = weight) %>%
naniar::miss_var_summary() %>%
dplyr::select(Variable = variable,
n = n_miss) %>%
flextable::flextable() %>%
apaSupp::theme_apa(caption = "Missing Data by Variable")Variable | n |
|---|---|
Weight, lbs | 11 |
IQ, pts | 8 |
Life Satisfaction | 1 |
Sex | 0 |
Age, yrs | 0 |
df_depress %>%
dplyr::select("Sex" = sex,
"Life Satisfaction" = lifesat,
"IQ, pts" = iq,
"Age, yrs" = age,
"Weight, lbs" = weight) %>%
naniar::gg_miss_upset()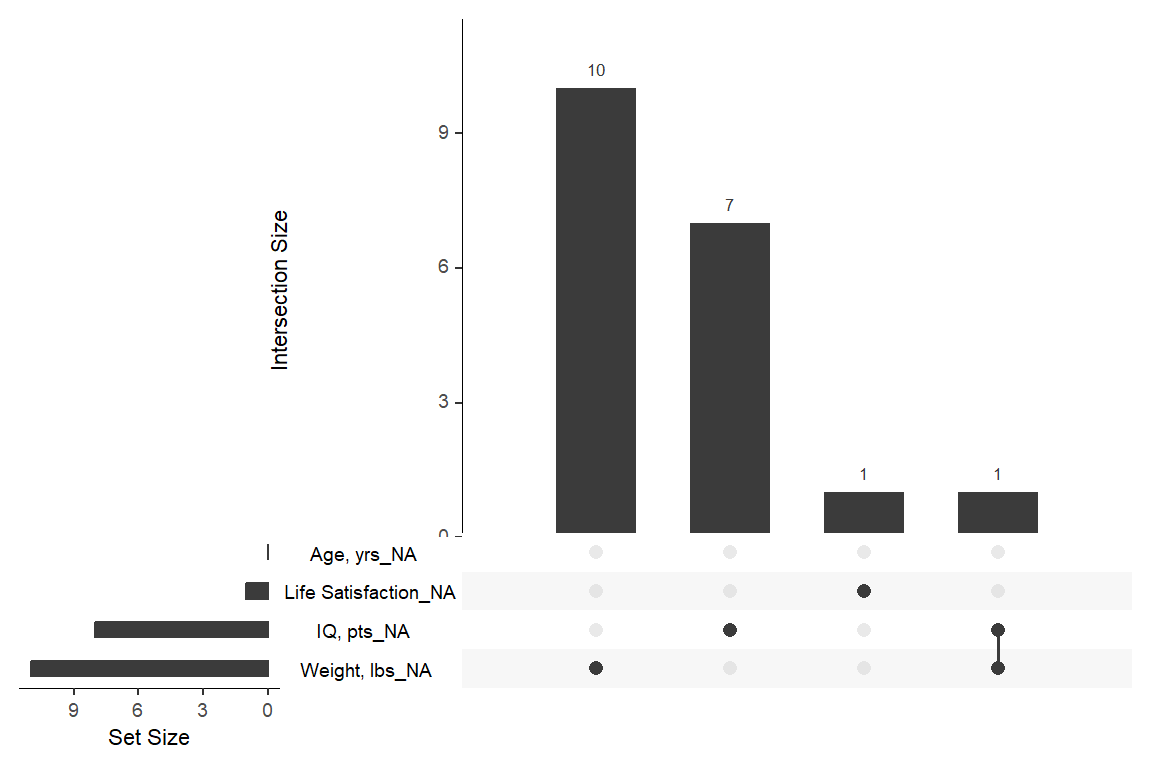
14.2.2 Summary
df_depress %>%
dplyr::select("Sex" = sex,
"Life Satisfaction" = lifesat) %>%
apaSupp::tab_freq(caption = "Descriptive Summary of Categorical Variables")Statistic | ||
|---|---|---|
Sex | ||
Male | 21 (17.8%) | |
Female | 97 (82.2%) | |
Life Satisfaction | ||
Yes (1) | 52 (44.1%) | |
No (0) | 65 (55.1%) | |
Missing | 1 (0.8%) | |
df_depress %>%
dplyr::select("IQ, pts" = iq,
"Age, yrs" = age,
"Weight, lbs" = weight) %>%
apaSupp::tab_desc(caption = "Descriptive Summary of Continuous Variables")NA | M | SD | min | Q1 | Mdn | Q3 | max | |
|---|---|---|---|---|---|---|---|---|
IQ, pts | 8 | 91.79 | 4.53 | 82.00 | 89.00 | 92.00 | 94.75 | 106.00 |
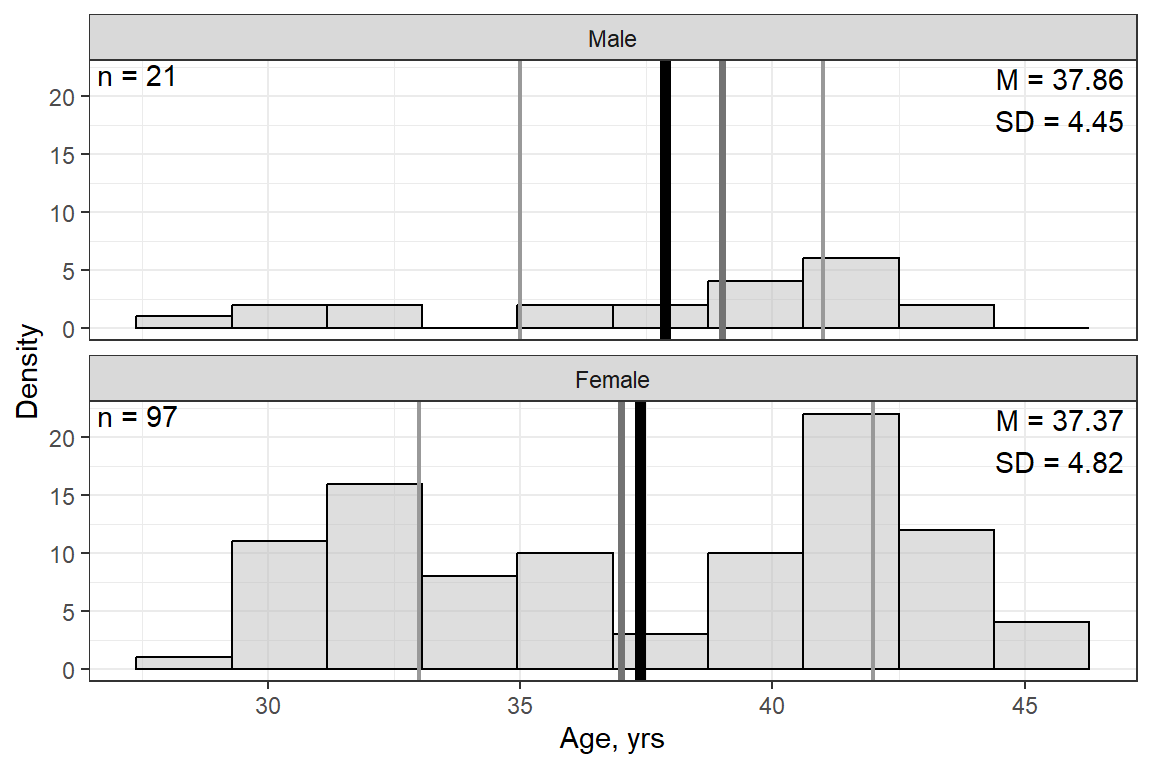
Age, yrs | 0 | 37.46 | 4.74 | 29.00 | 33.00 | 39.00 | 41.75 | 46.00 |
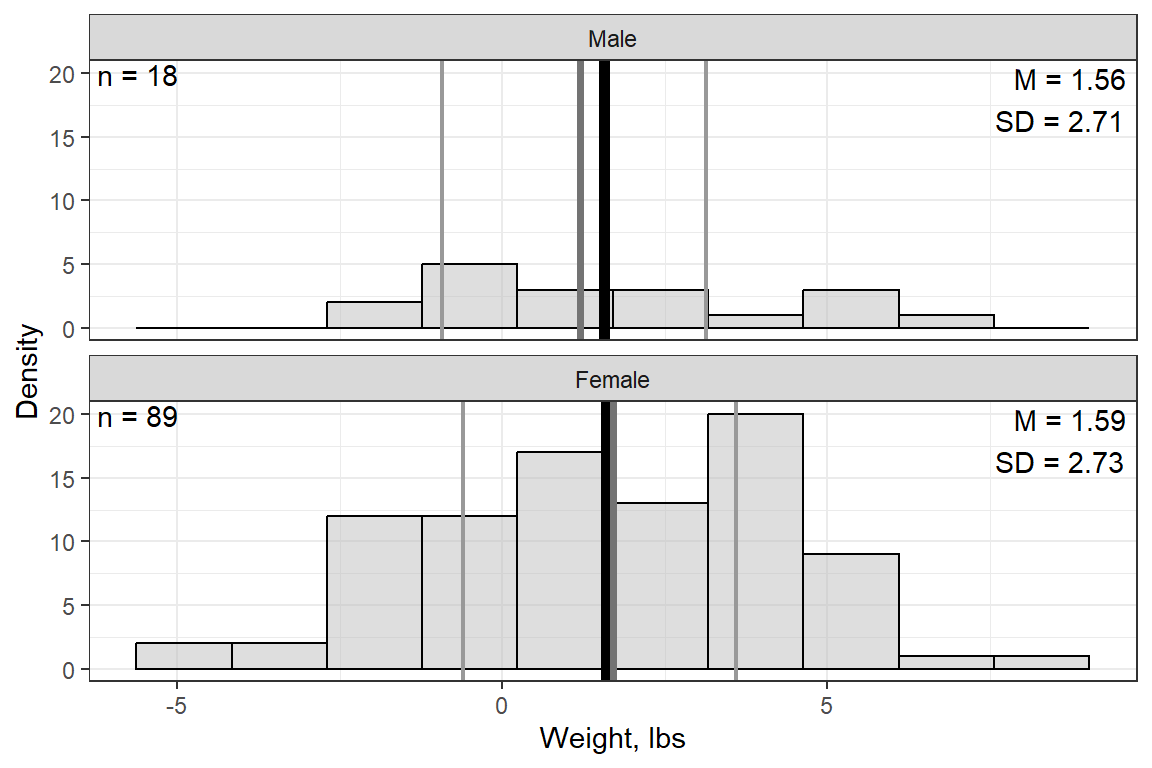
Weight, lbs | 11 | 1.59 | 2.72 | -4.90 | -0.65 | 1.70 | 3.55 | 8.30 |
Note. N = 118. NA = not available or missing; Mdn = median; Q1 = 25th percentile; Q3 = 75th percentile. | ||||||||
df_depress %>%
dplyr::select("Life Satisfaction" = lifesat,
"Sex" = sex,
"IQ" = iq,
"Age" = age,
"Weight" = weight) %>%
dplyr::mutate_all(as.numeric) %>%
apaSupp::tab_cor(caption = "Pairwise Correlations for Life Satisfaction, Sexd, IQ, Age, and Weight") %>%
flextable::hline(i = 4)Variable Pair | r | p | |
|---|---|---|---|
Life Satisfaction | Sex | .210 | .024* |
Life Satisfaction | IQ | .092 | .344 |
Life Satisfaction | Age | .100 | .269 |
Life Satisfaction | Weight | .059 | .550 |
Sex | IQ | .024 | .806 |
Sex | Age | -.039 | .672 |
Sex | Weight | .004 | .968 |
IQ | Age | -.430 | < .001*** |
IQ | Weight | -.290 | .003** |
Age | Weight | .420 | < .001*** |
Note. N = 118. r = Pearson's Product-Moment correlation coefficient. | |||
* p < .05. ** p < .01. *** p < .001. | |||
14.2.3 Visualize
df_depress %>%
dplyr::filter(complete.cases(lifesat, sex, iq, age, weight)) %>%
dplyr::select("Satisfaction" = lifesat,
"Sex" = sex,
"IQ, pts" = iq,
"Age, yrs" = age,
"Weight, lbs" = weight) %>%
GGally::ggpairs(aes(colour = Sex),
diag = list(continuous = GGally::wrap("densityDiag",
alpha = .3)),
lower = list(continuous = GGally::wrap("smooth",
shape = 16,
se = FALSE,
size = 0.75))) +
theme_bw() +
scale_fill_manual(values = c("blue", "coral")) +
scale_color_manual(values = c("blue", "coral")) 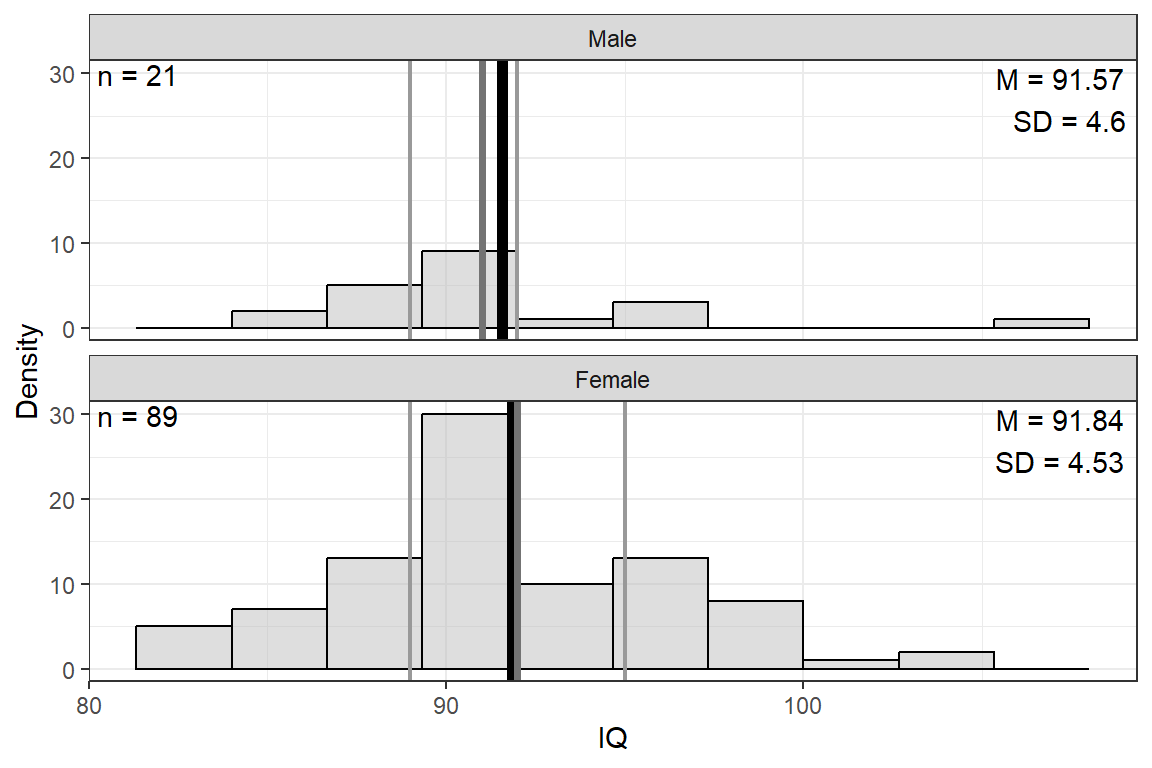
df_depress %>%
ggplot(aes(x = iq,
y = satlife)) +
geom_count() +
geom_smooth(method = "lm") +
theme_bw() +
labs(x = "IQ Score",
y = "Life Satisfaction, numeric") +
scale_y_continuous(breaks = 0:1,
labels = c("No (0)",
"Yes (1)"))
Figure 14.1
Hoffman’s Figure 2.3, top of page 46
df_depress %>%
ggplot(aes(x = weight,
y = satlife)) +
geom_count() +
geom_smooth(method = "lm") +
theme_bw() +
labs(x = "Weight, lbs",
y = "Life Satisfaction, numeric") +
scale_y_continuous(breaks = 0:1,
labels = c("No (0)",
"Yes (1)"))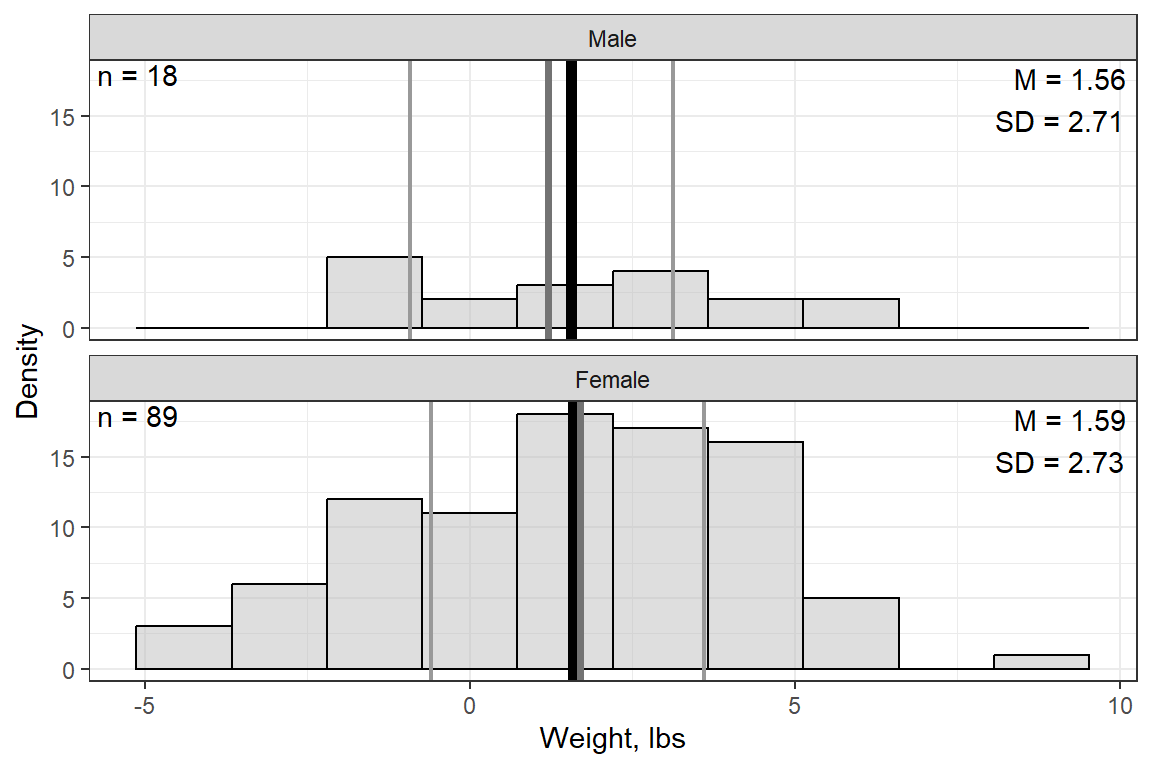
Hoffman’s Figure 2.3, top of page 46
df_depress %>%
ggplot(aes(x = age,
y = satlife)) +
geom_count() +
geom_smooth(method = "lm") +
theme_bw() +
labs(x = "Age in Years",
y = "Life Satisfaction, numeric") +
scale_y_continuous(breaks = 0:1,
labels = c("No (0)",
"Yes (1)"))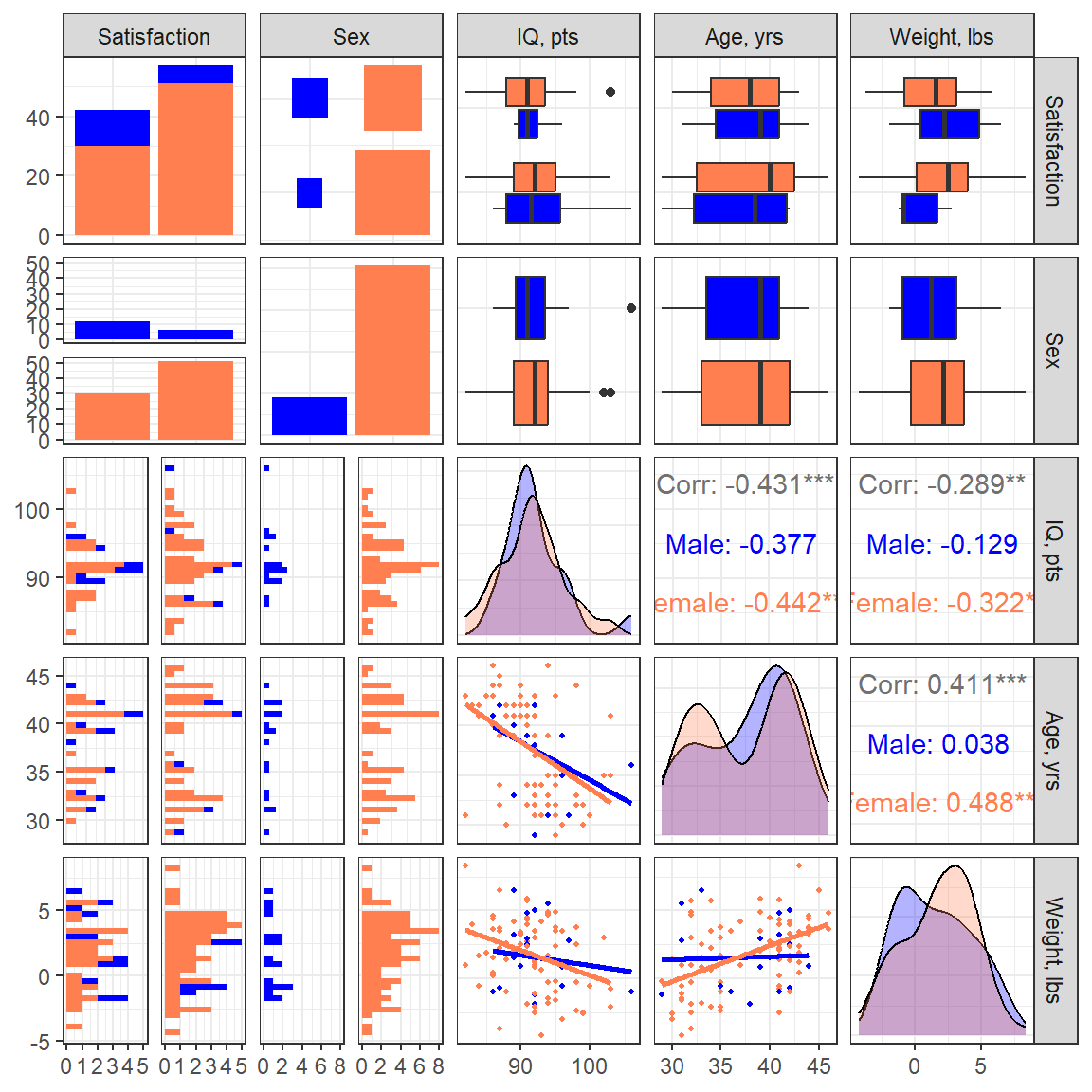
df_depress %>%
ggplot(aes(x = age,
y = satlife)) +
geom_count() +
geom_smooth(method = "lm") +
theme_bw() +
labs(x = "Age in Years",
y = "Life Satisfaction, numeric") +
facet_grid(~ sex) +
theme(legend.position = "none") +
scale_y_continuous(breaks = 0:1,
labels = c("No (0)",
"Yes (1)"))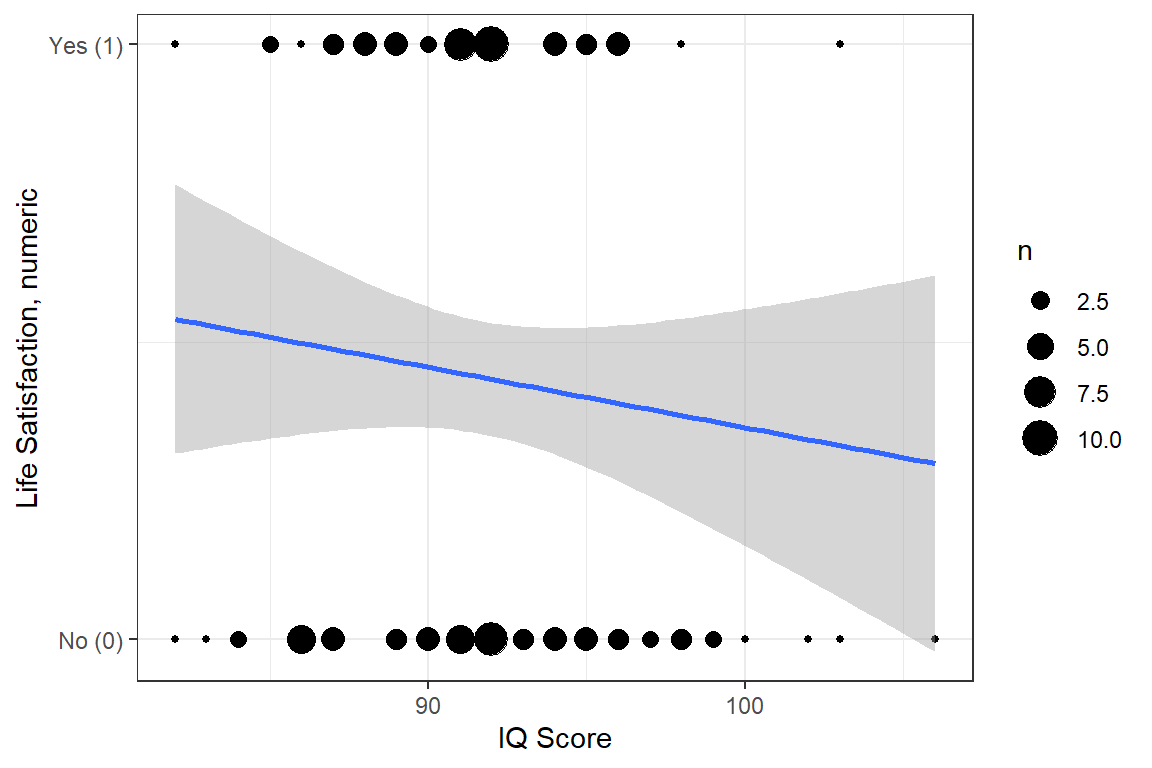
14.3 HAND CALCULATIONS
Probability, Odds, and Odds-Ratios
14.3.1 Marginal, over all the sample
Tally the number of participants who are satisfied vs. not…overall and by sex.
df_depress %>%
dplyr::select(sex,
"Satisfied with Life" = lifesat) %>%
apaSupp::tab_freq(split = "sex",
caption = "Observed Life Satisfaction by Sex")Male | Female | Total | ||||
|---|---|---|---|---|---|---|
Satisfied with Life | ||||||
Yes (1) | 14 (66.7%) | 38 (39.2%) | 52 (44.1%) | |||
No (0) | 7 (33.3%) | 58 (59.8%) | 65 (55.1%) | |||
Missing | 1 (1.0%) | 1 (0.8%) | ||||
14.3.2 Comparing by Sex
Cross-tabulate happiness (
satlife) withsex(male vs. female).
sex
satlife Male Female Sum
0 7 58 65
1 14 38 52
Sum 21 96 11714.3.2.1 Probability of being happy, by sex
Reference category = male
[1] 0.6666667Comparison Category = female
[1] 0.395833314.3.2.2 Odds of being happy, by sex
Reference category = male
[1] 2Comparison Category = female
[1] 0.655172414.3.2.3 Odds-Ratio for sex
\[ OR_{\text{female vs. male}} = \frac{odds_{female}}{odds_{male}} \]
[1] 0.3275862\[ OR_{\text{female vs. male}} = \frac{\frac{prob_{female}}{1 - prob_{female}}}{\frac{prob_{male}}{1 - prob_{male}}} \]
[1] 0.3275862\[ OR_{\text{female vs. male}} = \frac{\frac{n_{yes|female}}{n_{no|female}}}{\frac{n_{yes|male}}{n_{no|male}}} \]
[1] 0.327586214.4 SINGLE PREDICTOR
14.4.2 Parameter Table
Odds Ratio | Logit Scale | ||||||
|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | p | ||
sex | |||||||
Male | — | — | — | — | |||
Female | 0.33 | [0.11, 0.86] | < .0 | (0.51) | .028* | ||
(Intercept) | 0.69 | (0.46) | .134 | ||||
pseudo-R² | .044 | ||||||
Note. N = 117. CI = confidence interval. Significance denotes Wald t-tests for parameter estimates. Coefficient of determination displays Tjur's pseudo-R². | |||||||
* p < .05. ** p < .01. *** p < .001. | |||||||
apaSupp::tab_glm(fit_glm_1,
var_labels = c(sex = "Female vs. Male"),
pr2 = "both",
caption = "Parameter Etimates for Logistic Regressing for Life Satisfaction by Sex, Unadjusted Odds Ratio",
p_note = "apa1",
show_single_row = "sex")Odds Ratio | Logit Scale | ||||||
|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | p | ||
Female vs. Male | 0.33 | [0.11, 0.86] | -1.12 | (0.51) | .028* | ||
(Intercept) | 0.69 | (0.46) | .134 | ||||
pseudo-R² | |||||||
Tjur | .044 | ||||||
McFadden | .032 | ||||||
Note. N = 117. CI = confidence interval. Significance denotes Wald t-tests for parameter estimates. Coefficient of determination included for both Tjur and McFadden's pseudo-R². | |||||||
* p < .05. | |||||||
14.4.3 Model Fit
See this commentary by Paul Alison: https://statisticalhorizons.com/r2logistic/
# A tibble: 1 × 10
AIC AICc BIC R2_Tjur RMSE Sigma Log_loss Score_log Score_spherical PCP
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 160. 160. 165. 0.0437 0.486 1 0.665 -30.1 0.0550 0.528# R2 for Logistic Regression
Tjur's R2: 0.044# R2 for Generalized Linear Regression
R2: 0.032
adj. R2: 0.01914.4.4 Predicted Probability
14.4.4.1 Logit scale
sex emmean SE df asymp.LCL asymp.UCL
Male 0.693 0.463 Inf -0.214 1.6004
Female -0.423 0.209 Inf -0.832 -0.0138
Results are given on the logit (not the response) scale.
Confidence level used: 0.95 contrast estimate SE df z.ratio p.value
Male - Female 1.12 0.508 Inf 2.198 0.0280
Results are given on the log odds ratio (not the response) scale. 14.4.4.2 Response Scale
Probability
sex prob SE df asymp.LCL asymp.UCL
Male 0.667 0.1030 Inf 0.447 0.832
Female 0.396 0.0499 Inf 0.303 0.497
Confidence level used: 0.95
Intervals are back-transformed from the logit scale contrast odds.ratio SE df null z.ratio p.value
Male / Female 3.05 1.55 Inf 1 2.198 0.0280
Tests are performed on the log odds ratio scale 14.4.5 Interpretation
On average, two out of every three males is satisfied, b = 0.667, odds = 1.95, 95% CI [1.58, 2.40].
Females have 67% lower odds of being depressed, compared to men, b = -0.27, OR = 0.77, 95% CI [0.61, 0.96], p = .028.
14.4.6 Diagnostics
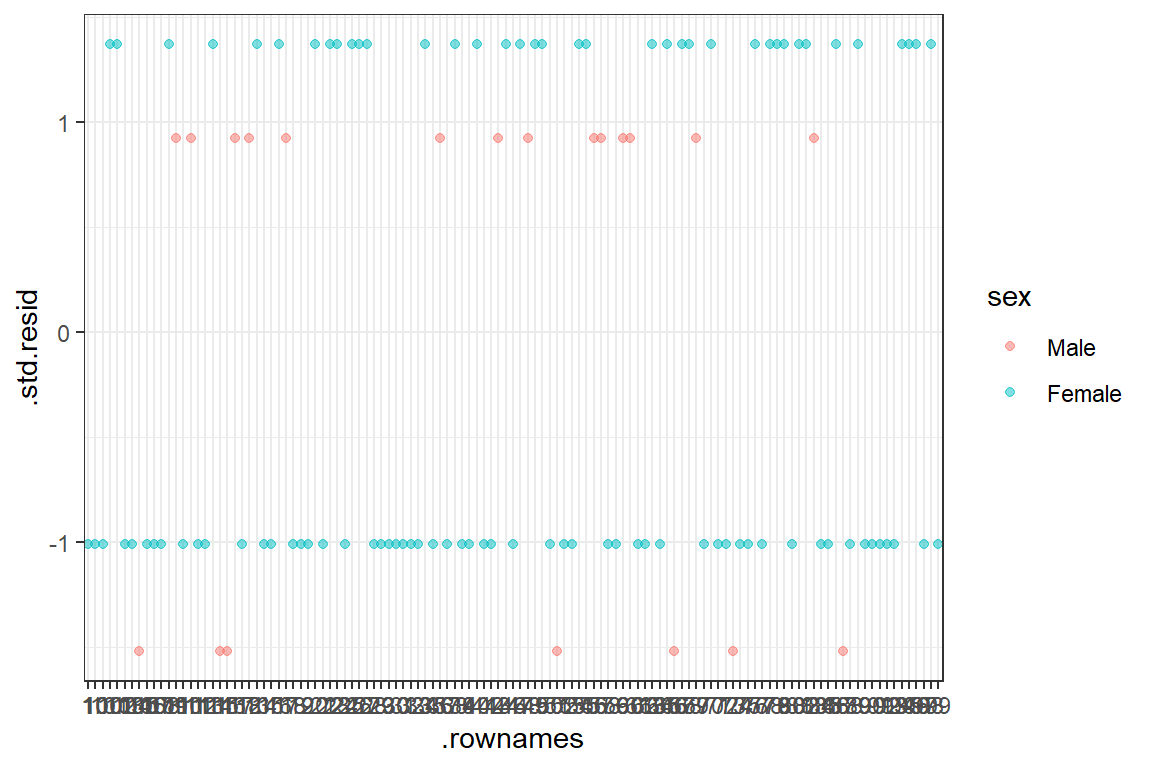
14.4.6.1 Influential values
Influential values are extreme individual data points that can alter the quality of the logistic regression model.
The most extreme values in the data can be examined by visualizing the Cook’s distance values. Here we label the top 7 largest values:
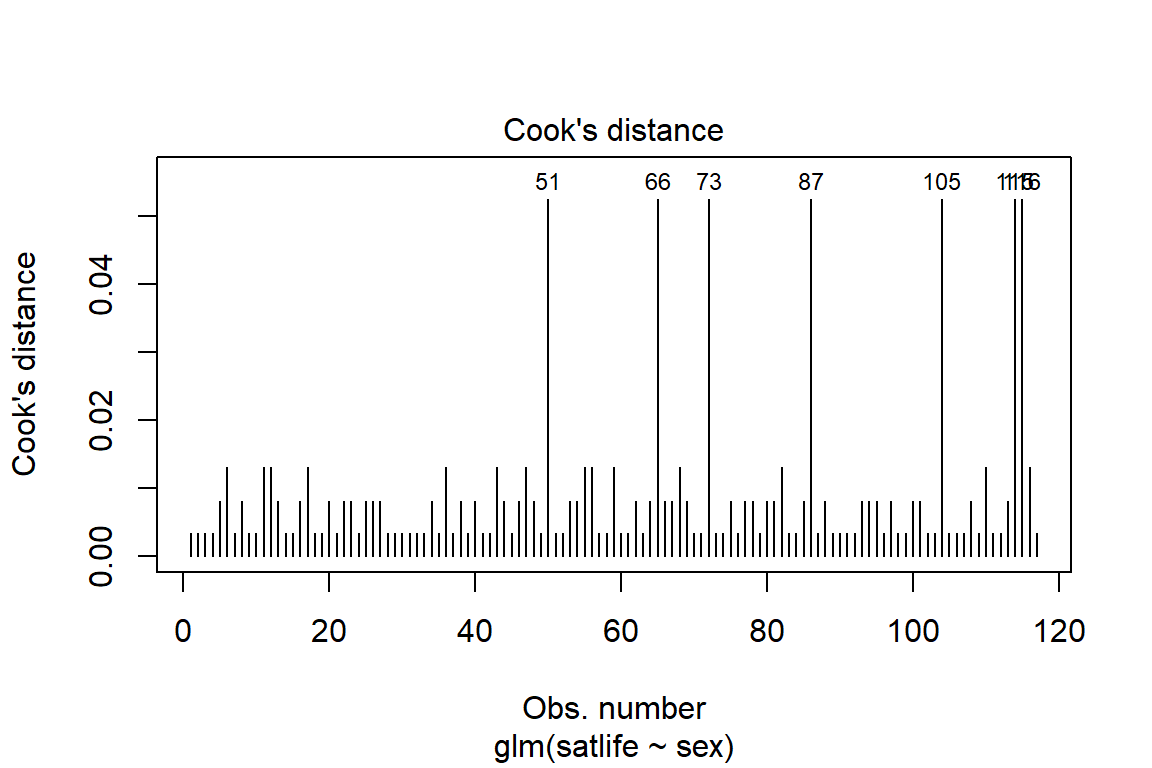
Note that, not all outliers are influential observations. To check whether the data contains potential influential observations, the standardized residual error can be inspected. Data points with an absolute standardized residuals above 3 represent possible outliers and may deserve closer attention.
14.5 MULTIPLE PREDICTORS
14.5.2 Parameter Table
EXAMPLE 3.3 A Logistic Regression Model of Life Satisfaction with Multiple Independent Variables, middle of page 52
Odds Ratio | Logit Scale | ||||||||
|---|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | Wald | LRT | VIF | ||
sex | .018* | 1.01 | |||||||
Male | — | — | — | — | |||||
Female | 0.28 | [0.09, 0.81] | < .0 | (0.56) | .022* | ||||
iq | 0.93 | [0.83, 1.03] | -0.07 | (0.05) | .174 | .167 | 1.29 | ||
age | 0.96 | [0.86, 1.06] | -0.04 | (0.05) | .433 | .431 | 1.42 | ||
weight | 0.96 | [0.81, 1.15] | -0.04 | (0.09) | .671 | .671 | 1.23 | ||
(Intercept) | 9.02 | (6.02) | .134 | ||||||
pseudo-R² | .075 | ||||||||
Note. N = 99. CI = confidence interval; VIF = variance inflation factor. Significance denotes Wald t-tests for individual parameter estimates, as well as Likelihood Ratio Tests (LRT) for single-predictor deletion. Coefficient of determination displays Tjur's pseudo-R². | |||||||||
* p < .05. ** p < .01. *** p < .001. | |||||||||
Odds Ratio | Logit Scale | |||||||
|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | Wald | LRT | ||
sex | .018* | |||||||
Male | — | — | — | — | ||||
Female | 0.28 | [0.09, 0.81] | < .0 | (0.56) | .022* | |||
iq | 0.93 | [0.83, 1.03] | -0.07 | (0.05) | .174 | .167 | ||
age | 0.96 | [0.86, 1.06] | -0.04 | (0.05) | .433 | .431 | ||
weight | 0.96 | [0.81, 1.15] | -0.04 | (0.09) | .671 | .671 | ||
(Intercept) | 9.02 | (6.02) | .134 | |||||
pseudo-R² | ||||||||
Tjur | .075 | |||||||
McFadden | .055 | |||||||
Note. N = 99. CI = confidence interval. Significance denotes Wald t-tests for individual parameter estimates, as well as Likelihood Ratio Tests (LRT) for single-predictor deletion. Coefficient of determination included for both Tjur and McFadden's pseudo-R². | ||||||||
* p < .05. ** p < .01. *** p < .001. | ||||||||
Odds Ratio | Logit Scale | ||||||||
|---|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | Wald | LRT | VIF | ||
sex | .018* | 1.01 | |||||||
Male | — | — | — | — | |||||
Female | 0.28 | [0.09, 0.81] | < .0 | (0.56) | .022* | ||||
iq | 0.93 | [0.83, 1.03] | -0.07 | (0.05) | .174 | .167 | 1.29 | ||
age | 0.96 | [0.86, 1.06] | -0.04 | (0.05) | .433 | .431 | 1.42 | ||
weight | 0.96 | [0.81, 1.15] | -0.04 | (0.09) | .671 | .671 | 1.23 | ||
AIC | 138 | ||||||||
BIC | 150 | ||||||||
(Intercept) | 9.02 | (6.02) | .134 | ||||||
pseudo-R² | .075 | ||||||||
Note. N = 99. CI = confidence interval; VIF = variance inflation factor. Significance denotes Wald t-tests for individual parameter estimates, as well as Likelihood Ratio Tests (LRT) for single-predictor deletion. Coefficient of determination displays Tjur's pseudo-R². | |||||||||
* p < .05. ** p < .01. *** p < .001. | |||||||||
Odds Ratio | Logit Scale | ||||||
|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | p | ||
sex | |||||||
Male | — | — | — | — | |||
Female | 0.28 | [0.09, 0.81] | < .0 | (0.56) | .022* | ||
iq | 0.93 | [0.83, 1.03] | -0.07 | (0.05) | .174 | ||
age | 0.96 | [0.86, 1.06] | -0.04 | (0.05) | .433 | ||
weight | 0.96 | [0.81, 1.15] | -0.04 | (0.09) | .671 | ||
(Intercept) | 9.02 | (6.02) | .134 | ||||
pseudo-R² | .075 | ||||||
Note. N = 99. CI = confidence interval. Significance denotes Wald t-tests for parameter estimates. Coefficient of determination displays Tjur's pseudo-R². | |||||||
* p < .05. ** p < .01. *** p < .001. | |||||||
apaSupp::tab_glm(fit_glm_2,
var_labels = c(sex = "Female vs. Male",
iq = "IQ, pts",
age = "Age, yrs",
weight = "Weight, lbs"),
pr2 = "both",
show_single_row = "sex",
p_note = "apa1",
caption = "Parameter Etimates for Logistic Regressing for Life Satisfaction by Sex, Controlling fro IQ, Age, and Weight") %>%
flextable::bold(i = c(2))Odds Ratio | Logit Scale | ||||||||
|---|---|---|---|---|---|---|---|---|---|
Variable | OR | 95% CI | b | (SE) | Wald | LRT | VIF | ||
Female vs. Male | 0.28 | [0.09, 0.81] | -1.28 | (0.56) | .022* | .018* | 1.01 | ||
IQ, pts | 0.93 | [0.83, 1.03] | -0.07 | (0.05) | .174 | .167 | 1.29 | ||
Age, yrs | 0.96 | [0.86, 1.06] | -0.04 | (0.05) | .433 | .431 | 1.42 | ||
Weight, lbs | 0.96 | [0.81, 1.15] | -0.04 | (0.09) | .671 | .671 | 1.23 | ||
(Intercept) | 9.02 | (6.02) | .134 | ||||||
pseudo-R² | |||||||||
Tjur | .075 | ||||||||
McFadden | .055 | ||||||||
Note. N = 99. CI = confidence interval; VIF = variance inflation factor. Significance denotes Wald t-tests for individual parameter estimates, as well as Likelihood Ratio Tests (LRT) for single-predictor deletion. Coefficient of determination included for both Tjur and McFadden's pseudo-R². | |||||||||
* p < .05. | |||||||||
pseudo- | |||||||
|---|---|---|---|---|---|---|---|
Model | N | k | McFadden | Tjur | AIC | BIC | RMSE |
Univariate | 117 | 2 | .032 | .044 | 159.62 | 165.14 | 0.49 |
Multivariate | 99 | 5 | .055 | .075 | 137.52 | 150.50 | 0.47 |
Note. Models fit to different samples. k = number of parameters estimated in each model. Larger values indicated better performance. Smaller values indicated better performance for Akaike's Information Criteria (AIC), Bayesian information criteria (BIC), and Root Mean Squared Error (RMSE). | |||||||
14.6 COMPARE MODELS
14.6.1 Refit to Complete Cases
Restrict the data to only participant that have all four of these predictors.
Refit Model 1 with only participant complete on all the predictors.
14.6.2 Parameter Table
| Model 1 | Model 2 | ||||
|---|---|---|---|---|---|---|
Variable | OR | 95% CI | p | OR | 95% CI | p |
sex | ||||||
Male | — | — | — | — | ||
Female | 0.29 | [0.09, 0.84] | .026* | 0.28 | [0.09, 0.81] | .022* |
iq | 0.93 | [0.83, 1.03] | .174 | |||
age | 0.96 | [0.86, 1.06] | .433 | |||
weight | 0.96 | [0.81, 1.15] | .671 | |||
Fit Metrics | ||||||
AIC | 133.7 | 137.5 | ||||
BIC | 138.9 | 150.5 | ||||
pseudo-R² | ||||||
Tjur | .053 | .075 | ||||
McFadden | .039 | .055 | ||||
Note. N = 99. CI = confidence interval.Coefficient of determination estiamted with both Tjur and McFadden's psuedo R-squared | ||||||
* p < .05. ** p < .01. *** p < .001. | ||||||
apaSupp::tab_glms(list("Univariate" = fit_glm_1_redo,
"Multivariate" = fit_glm_2_redo),
var_labels = c(sex = "Sex",
iq = "IQ Score",
age = "Age, yrs",
weight = "Weight, lbs"),
fit = "AIC",
pr2 = "tjur",
narrow = TRUE) %>%
flextable::bold(i = 3)
| Univariate | Multivariate | ||
|---|---|---|---|---|
Variable | OR | 95% CI | OR | 95% CI |
Sex | ||||
Male | — | — | — | — |
Female | 0.29 | [0.09, 0.84]* | 0.28 | [0.09, 0.81]* |
IQ Score | 0.93 | [0.83, 1.03] | ||
Age, yrs | 0.96 | [0.86, 1.06] | ||
Weight, lbs | 0.96 | [0.81, 1.15] | ||
AIC | 133.70 | 137.52 | ||
pseudo-R² | .053 | .075 | ||
Note. N = 99. CI = confidence interval.Coefficient of determination estiamted by Tjur's psuedo R-squared | ||||
* p < .05. ** p < .01. *** p < .001. | ||||
14.6.3 Comparison Criteria
apaSupp::tab_glm_fits(list("Univariate, Initial" = fit_glm_1,
"Univariate, Restricted" = fit_glm_1_redo,
"Multivariate" = fit_glm_2_redo))pseudo- | |||||||
|---|---|---|---|---|---|---|---|
Model | N | k | McFadden | Tjur | AIC | BIC | RMSE |
Univariate, Initial | 117 | 2 | .032 | .044 | 159.62 | 165.14 | 0.49 |
Univariate, Restricted | 99 | 2 | .039 | .053 | 133.70 | 138.89 | 0.48 |
Multivariate | 99 | 5 | .055 | .075 | 137.52 | 150.50 | 0.47 |
Note. Models fit to different samples. k = number of parameters estimated in each model. Larger values indicated better performance. Smaller values indicated better performance for Akaike's Information Criteria (AIC), Bayesian information criteria (BIC), and Root Mean Squared Error (RMSE). | |||||||
# A tibble: 2 × 5
`Resid. Df` `Resid. Dev` Df Deviance `Pr(>Chi)`
<dbl> <dbl> <dbl> <dbl> <dbl>
1 97 130. NA NA NA
2 94 128. 3 2.18 0.53714.7 CHANGING REFERENCE CATEGORY
[1] "Male" "Female"df_depress_ref <- df_depress %>%
dplyr::mutate(male = sex %>% forcats::fct_relevel("Female", after = 0)) %>% dplyr::mutate(female = sex %>% forcats::fct_relevel("Male", after = 0))[1] "Female" "Male" [1] "Male" "Female"fit_glm_2_male <- glm(satlife ~ male + iq + age + weight,
data = df_depress_ref,
family = binomial(link = "logit"))
fit_glm_2_female <- glm(satlife ~ female + iq + age + weight,
data = df_depress_ref,
family = binomial(link = "logit"))apaSupp::tab_glms(list("Reference = Male" = fit_glm_2_female,
"Reference = Female" = fit_glm_2_male)) %>%
flextable::bold(i = c(3, 9))
| Reference = Male | Reference = Female | ||||
|---|---|---|---|---|---|---|
Variable | OR | 95% CI | p | OR | 95% CI | p |
female | ||||||
Male | — | — | ||||
Female | 0.28 | [0.09, 0.81] | .022* | |||
iq | 0.93 | [0.83, 1.03] | .174 | 0.93 | [0.83, 1.03] | .174 |
age | 0.96 | [0.86, 1.06] | .433 | 0.96 | [0.86, 1.06] | .433 |
weight | 0.96 | [0.81, 1.15] | .671 | 0.96 | [0.81, 1.15] | .671 |
male | ||||||
Female | — | — | ||||
Male | 3.59 | [1.24, 11.49] | .022* | |||
Fit Metrics | ||||||
AIC | 137.5 | 137.5 | ||||
BIC | 150.5 | 150.5 | ||||
pseudo-R² | ||||||
Tjur | .075 | .075 | ||||
McFadden | .055 | .055 | ||||
Note. N = 99. CI = confidence interval.Coefficient of determination estiamted with both Tjur and McFadden's psuedo R-squared | ||||||
* p < .05. ** p < .01. *** p < .001. | ||||||